

Interested in Quantitative or Physical Biology?Visit our Contact Us page for more information or to schedule a lab visit! |
|
 |
Bacterial ultrastructureOne of the most aesthetically appealing problems in biology is elucidating the mechanisms that give rise to spatial organization in biological systems. Far from being well-mixed, almost all biological systems exhibit precise spatial and temporal control of protein, mRNA, and DNA concentration, demonstrating that cells measure distance and detect proximity with a molecular-scale tool kit. Although these phenomena have traditionally been studied in the context of the detailed expression patterning in development, recent exciting results using high-throughput microscopy techniques (including our own work, Publications) reveal that precise spatial organization is the rule rather than the exception in the bacterial cell. |
 |
Chromosome structure and dynamicsThe mechanisms by which the prokaryotic cell organizes and segregates its chromosome remain largely unknown in spite of a long standing appreciation of the importance of these processes. The Wiggins lab has been investigating the physical structure, organization, and dynamics of the prokaryotic chromosome by exploiting three complementary approaches: live-cell imaging, in vitro single-molecule experiments investigating proteins-DNA interactions, and biophysical modeling. |
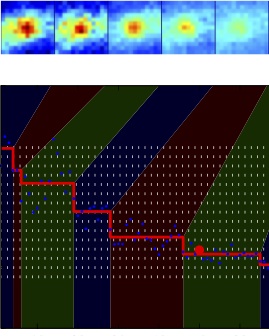 |
Resolving DNA replication conflicts during the cell cycleOne of the most important biological processes for the cell is faithful replicaiton of its genetic material prior to division. During this dynamic process the replication machinery is thought to encounter many conflicts, such as DNA-bound protein or transcription machinery, that lead to replication collisions or stalls. Although ubiquiutous, a mechanistic understanding of what happens to the replication machinery during one of these conflicts is still a mystery. Collaborating with the Merrikh Lab in UW Microbiology, we use a single-molecule approach to attempt to visualize and characterize these conflicts in living cells. |
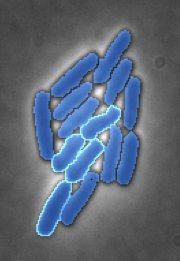 |
Bacterial interactions & cooperativityAlthough usually considered relatively simple single-celled organisms, bacteria have developted a number of complex mechanisms to interact with their neighbors. Many of these systems are thought to be triggered by physical mechanisms, such as cell-to-cell contact or surface sensing, but the biophysical processes that are responsible for this regulation remain largely a mystery. We are working in collaboration with the Mougous Lab in UW Microbiology to quantitatively characterize the dynamics of one of these inter-cellular systems, the Type VI secretion system. |
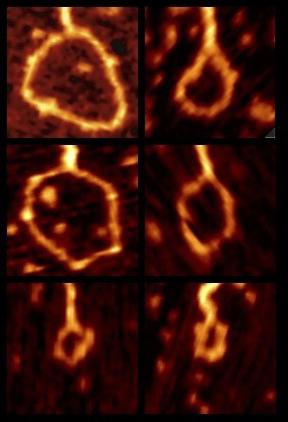 |
DNA chain statistics for biological applicationsWhat role does DNA statistics play in transcriptional regulation? Given the genomic locations of gene regulatory sequences relative to the promoter, what is their relative importance? (See description above.) The Wiggins lab is currently working on practical computational tools for biologist to help answer these questions quantitatively. |
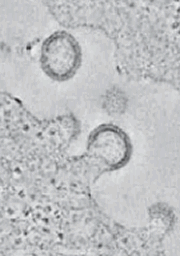 |
Membranes, geometry & force:We have proposed that in some biological contexts it is possible to deduce the forces applied to a membrane from its conformation, captured via cryo electron microscopy tomograms (three dimensional reconstructions). We are currently experimenting with this new technique in in vitro experiments. |
Lab MicroscopesFor more information on using the microscopes or for collaboration requests, please contact a lab member. |
|
 |
"Super-Scope"is a home-brew single-molecule super-resolution scope with TIRF and dual-view modes. The microscope is controlled using the Micro-Manager software package. It has autofocus capabilities, an Andor EMCCD camera, and four excitation sources: 566nm, 514nm, 488nm, 405nm. |
"Octoscope"is a commerical Nikon Eclipse Ti, with an arc-lamp, environmental chamber, perfect focus, a motorized stage, and imaging using either the Coolsnap HQ2 or K4 camera. This scope currently has filter sets for mCherry, dsRed, YFP, GFP, CFP, and DAPI and is used principly for multi-hour timelapse and high-throughput imaging. We use the Elements software package to control this scope. Objectives: 100x Ph3 1.4 NA, 60x Ph3 1.4 NA. |  |
 |
"Zeiss"is an old Axioplan II Zeiss scope controlled by Micro-Manager. This scope uses a Photometrics FX cooled CCD camera, 100x Phase3 objective, and has a motorized z-drive, objective turret, and filter wheel. |