| Home | Gene Index | Wiggins Lab | Invert Page |
"pyruvate dehydrogenase (pyruvate oxidase), thiamin-dependent, FAD-binding"
Gene Ontology: | |
| GO:0000287 | magnesium ion binding |
| GO:0003824 | catalytic activity |
| GO:0005829 | cytosol |
| GO:0005886 | plasma membrane |
| GO:0008289 | lipid binding |
| GO:0016020 | membrane |
| GO:0016491 | oxidoreductase activity |
| GO:0030976 | thiamine pyrophosphate binding |
| GO:0052737 | pyruvate dehydrogenase (quinone) activity |
| GO:0055114 | oxidation-reduction process |
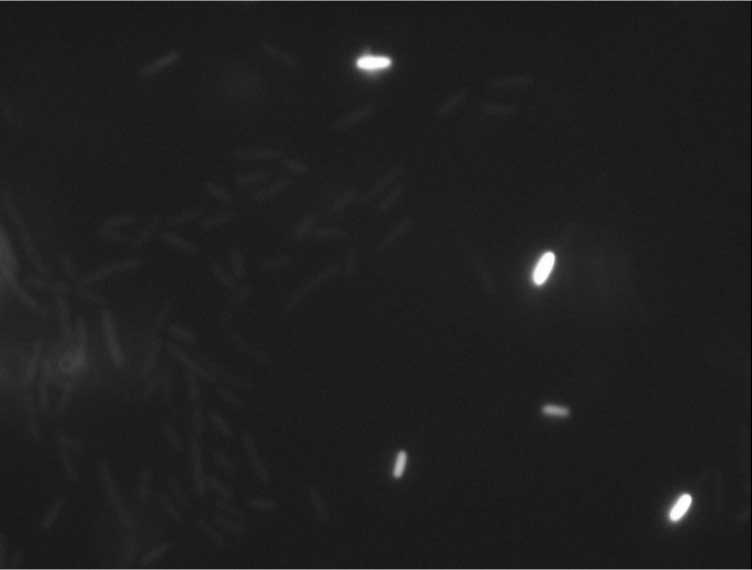
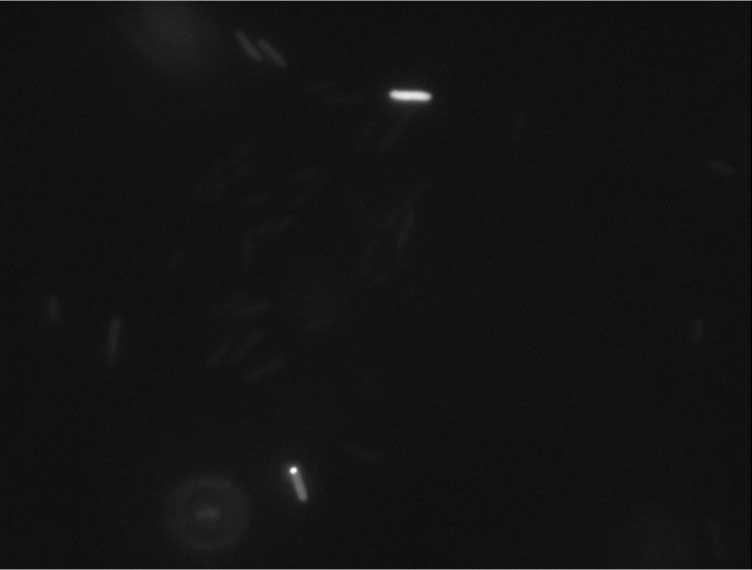
Protein localization Map: poxB | |||
| Gene name: | poxB | ||
| Strain: | JW0855 | ||
| Induction: | 50 uM IPTG | ||
| Comment: | 4A6C | ||
| Reference: | PMID:16769691--ASKA Collection--Kitagawa et al. | ||
|
Cell Orientation
|
Relative Intensity
|
|
Most similar localization patterns. gene: distance | ||||
|
|
|
|
|
|
|
|
|
|