Tutorial for cell file manipulations
This tutorial shows some examples on how to use the cell files for your analysis.
Contents
Get location of the directory and the cell file folder
dirname = [filepath,'60mrnaCropped']; cellFileFolder = [dirname, filesep,'xy1/cell'];
Load cell with ID 11
currentCell = load([cellFileFolder,filesep,'cell0000011.mat']); % display the information in the cell file currentCell % print the motherID disp (['mother id : ', num2str(currentCell.motherID)]); % display the information at 12 frames after birth disp('Cell Information at frame 12'); currentCell.CellA{12} % show the foci scores at 12 frames after birth disp('Foci scores at frame 12'); currentCell.CellA{12}.locus1(:).score % information for the first focus disp('First focus'); currentCell.CellA{12}.locus1(1) % information for the second focus disp('Second focus'); currentCell.CellA{12}.locus1(2)
currentCell =
CellA: {1x37 cell}
death: 37
birth: 1
divide: 1
sisterID: 0
motherID: 0
daughterID: [27 28]
ID: 11
neighbors: []
stat0: 0
ehist: 0
contactHist: 0
mother id : 0
Cell Information at frame 12
ans =
cellLength: [32 8.7198]
xx: [1x41 double]
yy: [1x23 double]
mask: [23x41 logical]
r_offset: [266 217]
BB: [266 217 40 22]
edgeFlag: 0
contactHist: 0
numNeighbors: 0
phase: [23x41 uint16]
coord: [1x1 struct]
length: [32 13]
pole: [1x1 struct]
fluor1: [23x41 uint16]
fluor1mm: [358 3610]
fl1: [1x1 struct]
cell_dist: 1
gray: 5.4220e+03
locus1: [1x2 struct]
fluor1_filtered: [23x41 double]
r: [285.8996 227.6022]
error: [1x1 struct]
ehist: 0
stat0: 0
Foci scores at frame 12
ans =
6.9524
ans =
3.9377
First focus
ans =
r: [298.8971 227.7778]
score: 6.9524
intensity: 11.0691
normIntensity: 11.0691
shortaxis: -1.9693
longaxis: 12.8486
fitSigma: 0.8483
fitScore: 0.9444
Second focus
ans =
r: [283.5400 222.8604]
score: 3.9377
intensity: 6.1612
normIntensity: 6.1612
shortaxis: -4.2879
longaxis: -3.1091
fitSigma: 0.8401
fitScore: 0.9517
Foci per frame
Make an array of number of foci per frame with score larger than 3 for this cell
frames = numel(currentCell.CellA); fociPerFrame = zeros(1,frames); for t = 1 : frames numFoci = numel(currentCell.CellA{t}.locus1); for i = 1 : numFoci if (currentCell.CellA{t}.locus1(i).score > 3) fociPerFrame(t) = fociPerFrame(t) + 1; end end end fociPerFrame
fociPerFrame =
Columns 1 through 13
1 1 1 1 1 1 2 2 1 1 1 2 1
Columns 14 through 26
1 1 1 2 2 1 2 1 2 1 1 0 3
Columns 27 through 37
3 2 2 2 2 2 2 2 1 1 2
Get cell files
allcells = dir([cellFileFolder,filesep '*ell*.mat']) CompleteCellCylceCells = dir([cellFileFolder,filesep 'Cell*.mat'])
allcells =
44x1 struct array with fields:
name
date
bytes
isdir
datenum
CompleteCellCylceCells =
4x1 struct array with fields:
name
date
bytes
isdir
datenum
Count cells
Go through every cell file and count the number cells with at least one foci at birth above score 3
count = 0; for i = 1 : numel(allcells) cellname = allcells(i).name; currentCell = load([cellFileFolder,filesep,cellname]); birth = 1; if isfield(currentCell.CellA{birth},'locus1') && currentCell.CellA{birth}.locus1(1).score>3 count = count + 1; end end disp (['Cells with at least one foci with score > 3 : ', num2str(count)]);
Cells with at least one foci with score > 3 : 21
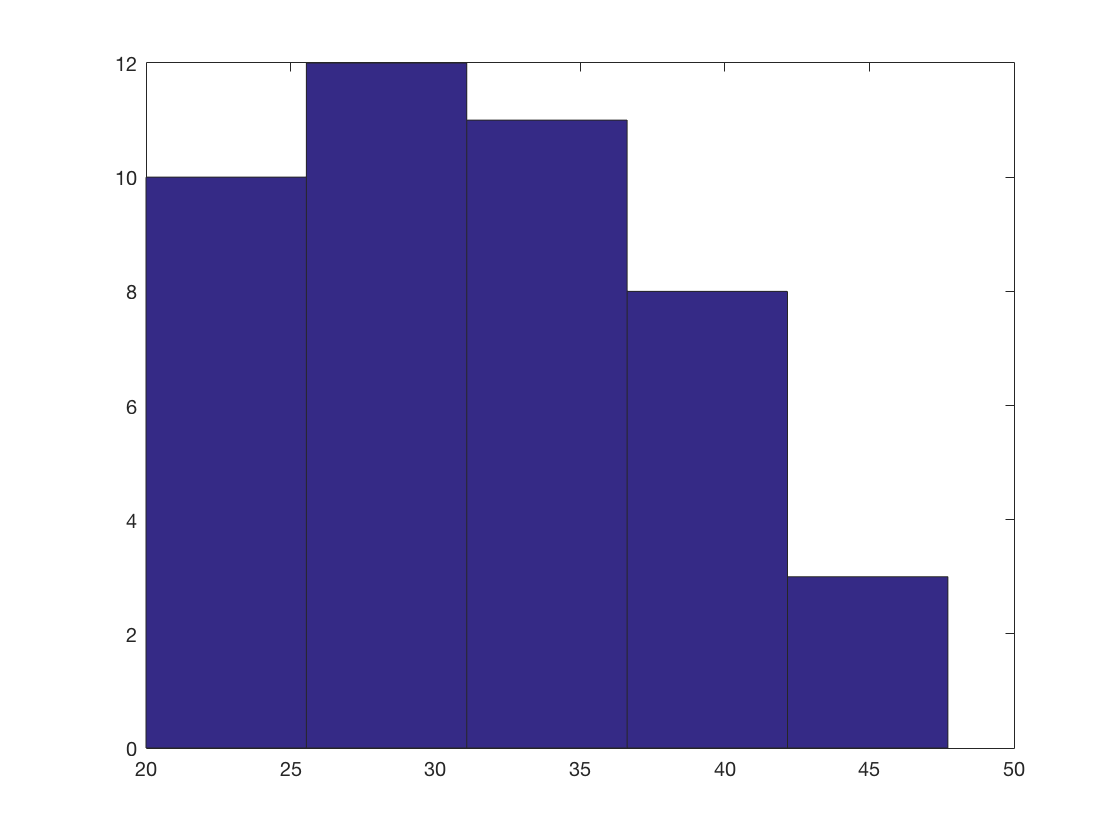
Average Cell Length
Go through every cell file, calculate the mean cell length, and make a histogram
% array of the average length for each cell averageCellLengths = []; for i = 1 : numel(allcells) cellname = allcells(i).name; currentCell = load([cellFileFolder,filesep,cellname]); % drill is a function that extracts a quantity from cellA for all time % frames lengthPerFrame = drill (currentCell.CellA, '.cellLength(1)'); avCellLength = mean(lengthPerFrame); averageCellLengths(i) = avCellLength; end hist(averageCellLengths,5)