| Home | Gene Index | Wiggins Lab | Invert Page |
predicted DNA-binding transcriptional regulator
Gene Ontology: | |
| GO:0000160 | phosphorelay signal transduction system |
| GO:0000166 | nucleotide binding |
| GO:0003677 | DNA binding |
| GO:0003700 | sequence-specific DNA binding transcription factor activity |
| GO:0004871 | signal transducer activity |
| GO:0005524 | ATP binding |
| GO:0006351 | transcription, DNA-dependent |
| GO:0006355 | regulation of transcription, DNA-dependent |
| GO:0007165 | signal transduction |
| GO:0008134 | transcription factor binding |
| GO:0017111 | nucleoside-triphosphatase activity |
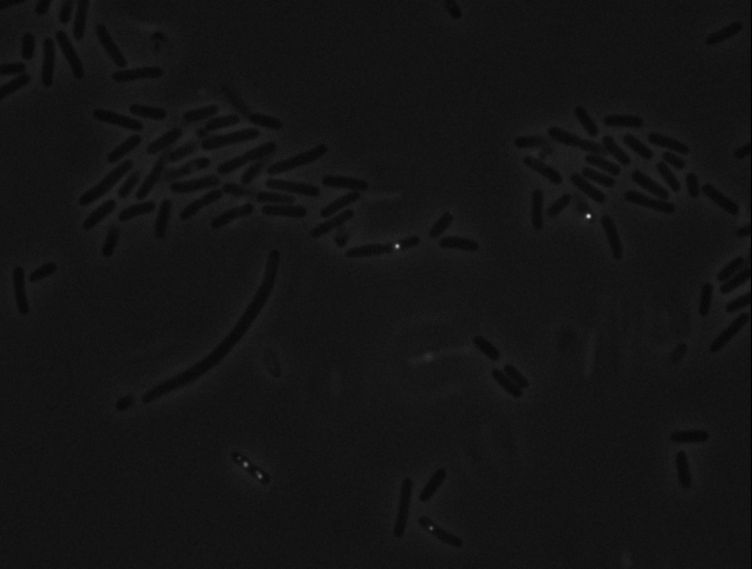
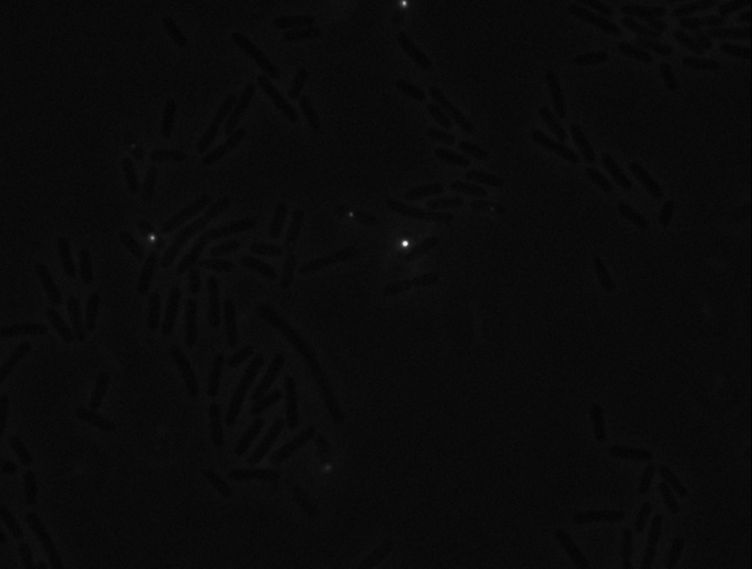
Protein localization Map: ygeV | |||||
| Gene name: | ygeV | Gene name: | ygeV | ||
| Strain: | JW2837 | Strain: | JW2837 | ||
| Induction: | 500 uM IPTG | Induction: | 50 uM IPTG | ||
| Comment: | 6A4H | Comment: | 6A4H | ||
| Reference: | PMID:16769691--ASKA Collection--Kitagawa et al. | Reference: | PMID:16769691--ASKA Collection--Kitagawa et al. | ||
|
|
Cell Orientation
|
Relative Intensity
|
||
Most similar localization patterns. gene: distance | ||||
|
|
|
|
|
|
|
|
|
|