| Home | Gene Index | Wiggins Lab | Invert Page |
fused DNA-binding transcriptional regulator, proline dehydrogenase, pyrroline-5-carboxylate dehydrogenase
Gene Ontology: | |
| GO:0000986 | bacterial-type RNA polymerase core promoter proximal region sequence-specific DNA binding |
| GO:0001141 | bacterial-type RNA polymerase core promoter proximal region sequence-specific DNA binding transcription factor activity involved in negative regulation of transcription |
| GO:0003677 | DNA binding |
| GO:0003842 | 1-pyrroline-5-carboxylate dehydrogenase activity |
| GO:0004657 | proline dehydrogenase activity |
| GO:0006351 | transcription, DNA-dependent |
| GO:0006355 | regulation of transcription, DNA-dependent |
| GO:0006537 | glutamate biosynthetic process |
| GO:0006561 | proline biosynthetic process |
| GO:0009898 | internal side of plasma membrane |
| GO:0010133 | proline catabolic process to glutamate |
| GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor |
| GO:0050660 | flavin adenine dinucleotide binding |
| GO:0055114 | oxidation-reduction process |
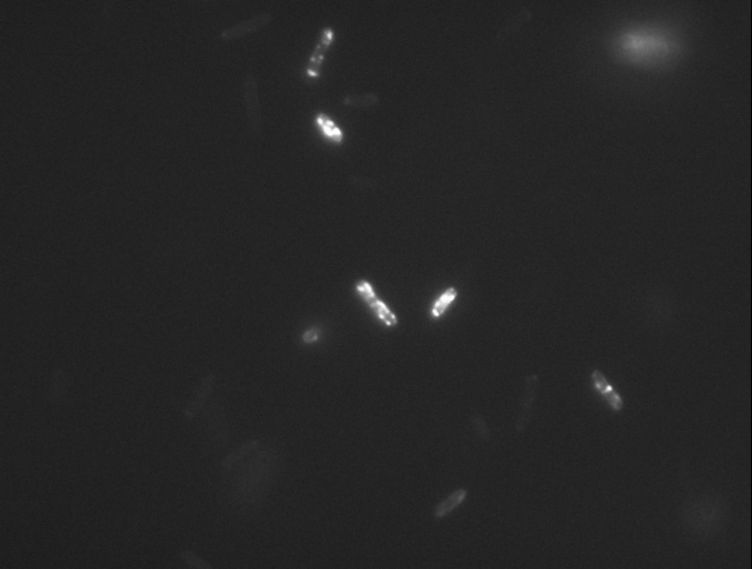
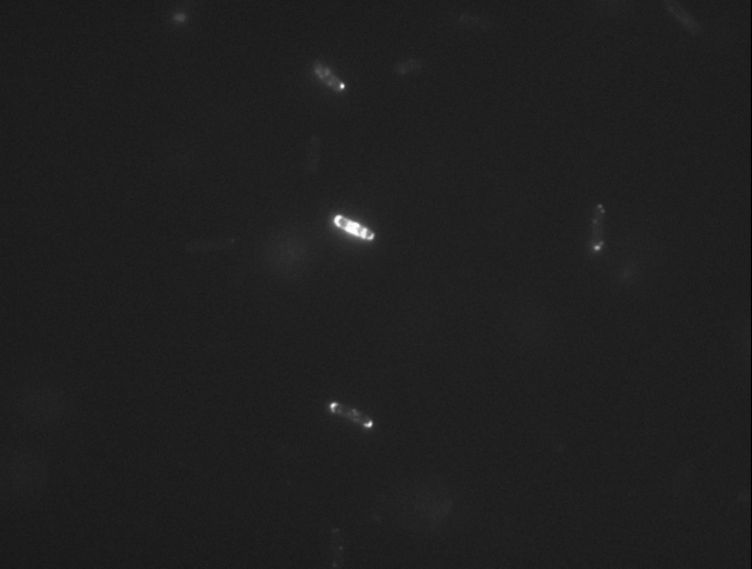
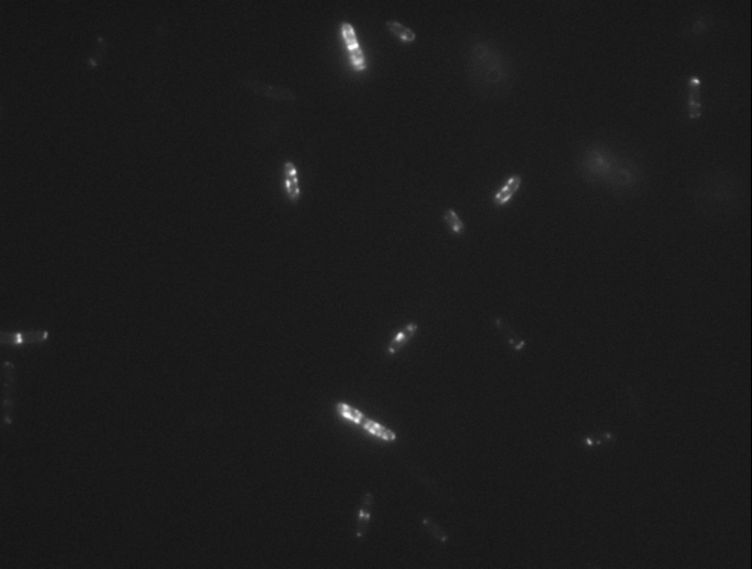
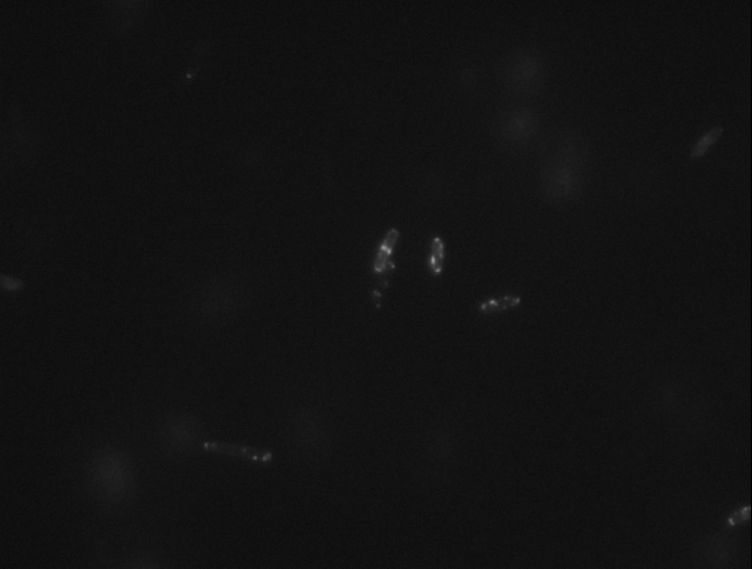
Protein localization Map: putA | |||||
| Gene name: | putA | Gene name: | putA | ||
| Strain: | JW0999 | Strain: | JW0999 | ||
| Induction: | 500 uM IPTG | Induction: | 50 uM IPTG | ||
| Comment: | 4B8D | Comment: | 4B8D | ||
| Reference: | PMID:16769691--ASKA Collection--Kitagawa et al. | Reference: | PMID:16769691--ASKA Collection--Kitagawa et al. | ||
|
|
Cell Orientation
|
Relative Intensity
|
||
Most similar localization patterns. gene: distance | ||||
|
|
|
|
|
|
|
|
|
|